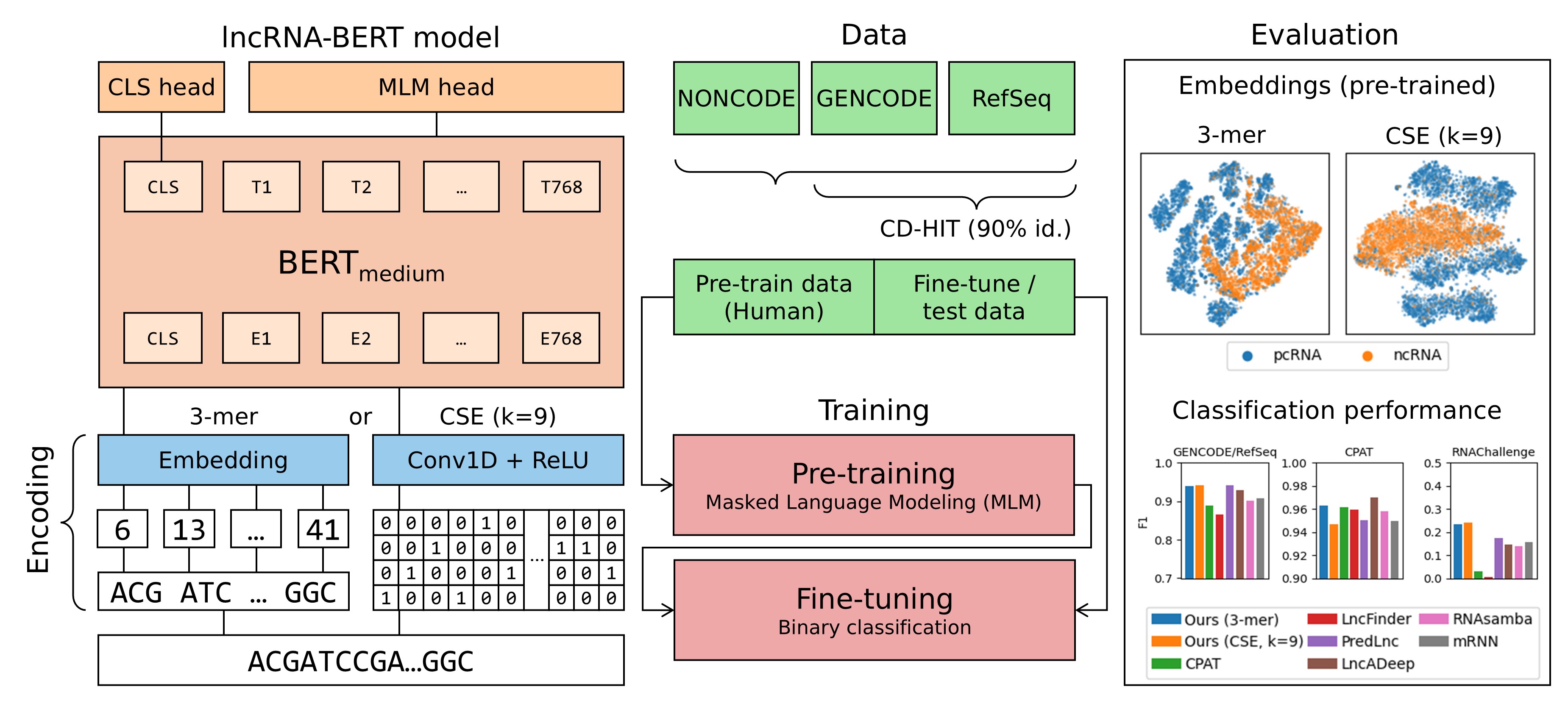
Experiments
This page contains descriptions of experiments that were conducted during the development of lncRNA-BERT, in addition to the experiments reported in LncRNA-BERT: An RNA Language Model for Classifying Coding and Long Non-Coding RNA.
Main Experiments
In our paper, we carry out
experiments that compare different pre-training data configurations as well as
different encoding methods. Scripts to reproduce these experiments can be found
in the experiments folder in
our repository. This folder also contains scripts for training the final
lncRNA-BERT models (with 3-mer tokenization or Convolutional Sequence Encoding).
Note that each of these bash scripts call upon the regular Python scripts that
are part of the lncrnapy package.
Hyperparameter Tuning
Hyperparameter tuning was carried out at several stages during the development of lncRNA-BERT.
Here, we aim to provide a clear overview of the settings that were experimented with and the decisions that were made.
The results are subject to improvements that were made in lncRNA-BERT’s implementation, as well as minor differences datasets utilized throughout development.
The described experiments below are targeted towards allowing a fair comparisons between pre-training data, encoding methods, and other NLMs, as reported in our paper.
Nevertheless, we acknowledge that lncRNA-BERT’s performance can likely be improved with additional tuning.
We report hyperparameters with their appropriate names, such that experiments can be repeated by using lncRNA-Py’s scripts.
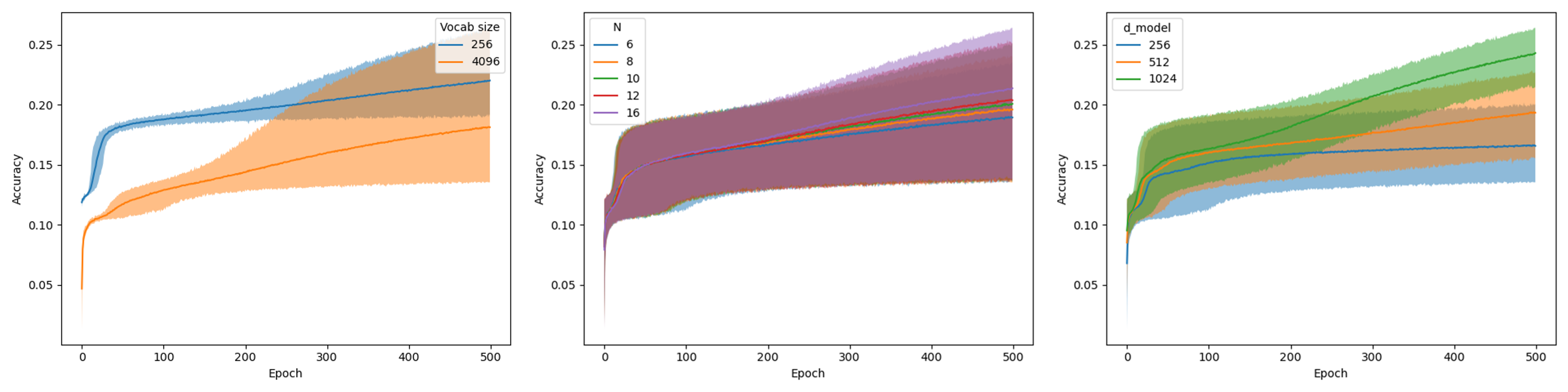
BERT configuration
We experimented with the following architecture parameters of BERT, for pre-training, using BPE with vocab_size 256 and 4096:
Transformer blocks (
N): 6, 8, 10, 12, 16Dimensionality (
d_model): 256, 512, 1024
In total, this yields 2*5*3=30 configurations. The graphs in the visualizations below represent the average for each hyperparameter value (see legend), while the bands around the graphs represent the minimum and maximum performance for all settings, given that specific hyperparameter value.
Larger values for d_model and N lead to increased MLM accuracy.
The figure indicates that for vocab_size, 4096 requires a larger model than 256.
Moreover, d_model is shown to be more impactful than N.
Ultimately, we chose to adopt BERT-medium’s settings, mainly to ensure a fair comparison to methods like DNABERT-2, BiRNA-BERT, and GENA-LM, which use a similar configuration.
This configuration uses a relatively large dimensionality (d_model=768), which was shown to be beneficial, while setting its number of transformer blocks to a not-so-extreme value (N=12).
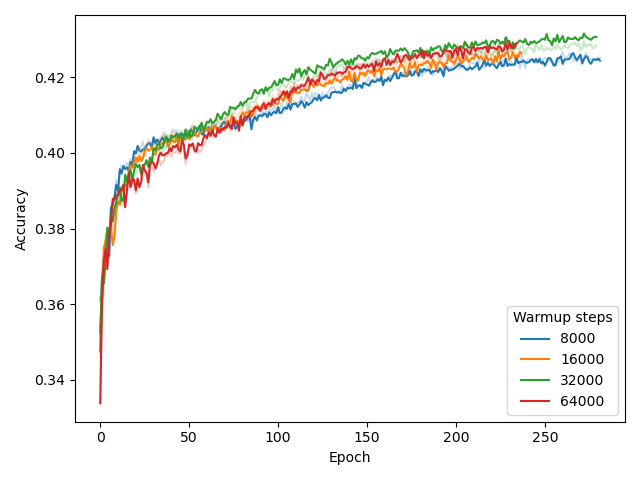
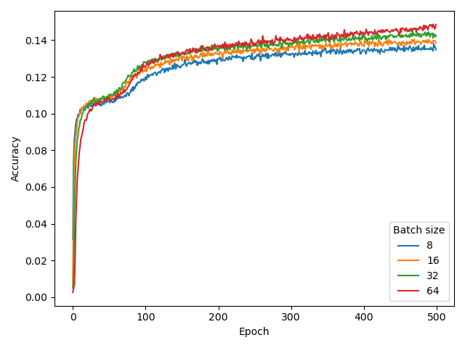
Masked Language Modeling
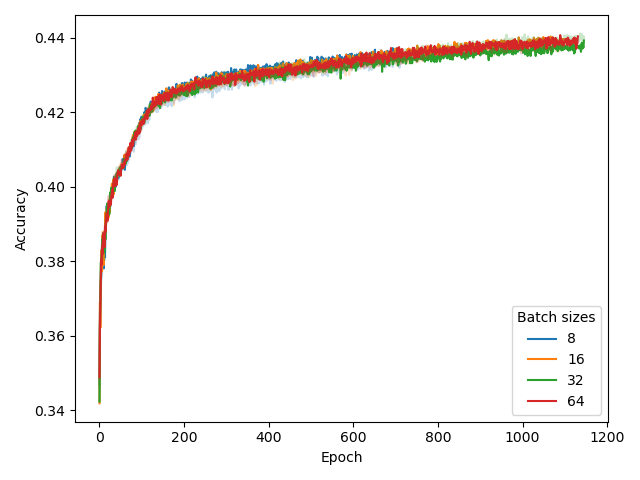
For MLM, we set warmup_steps=32000 and batch_size=8. The experiments that led to this conclusion are reported here, using model (N=6, d_model=256) and BPE with vocab_size=4096.
MLM accuracy (% of correctly predicted masked tokens) is reported on the y-axis.
While our choice for warmup_steps (while using batch_size=8) is obvious from the figure , deciding batch_size=8 was motivated by a previous experiment as well as compute limitations, rather than these results.
Specifically, our BERT configuration experiment pointed out that using a larger model is beneficial for performance, much more beneficial than the effect of batch_size, shown above.
Unfortunately, using large models in combinations with large batch sizes is computationally demanding, as it requires significant GPU memory.
Hence, we chose to prioritize model size over batch size here, setting batch_size=8 to save computational expenses for other experiments.
We repeated the experiment for a larger model (N=12, d_model=1024) with CSE in a much later development stage, confirming that batch_size still only had limited effect.
Classification
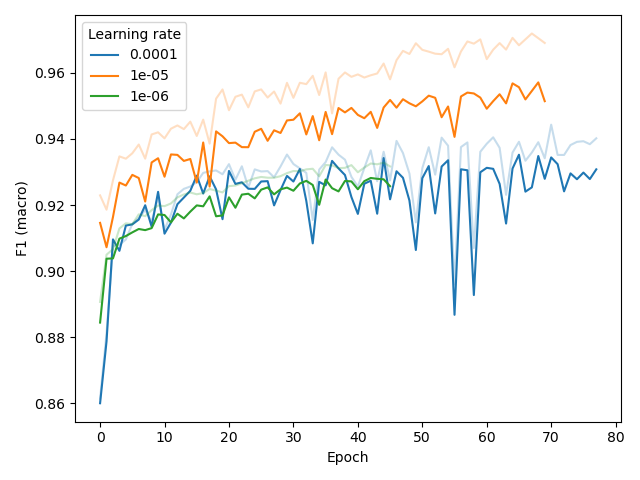
For the fine-tuning task, we initially set learning rate (lr) learning_rate=1e-05 based on an experiment (below, left) with BPE vocab_size=4096, and later confirmed that this setting was also
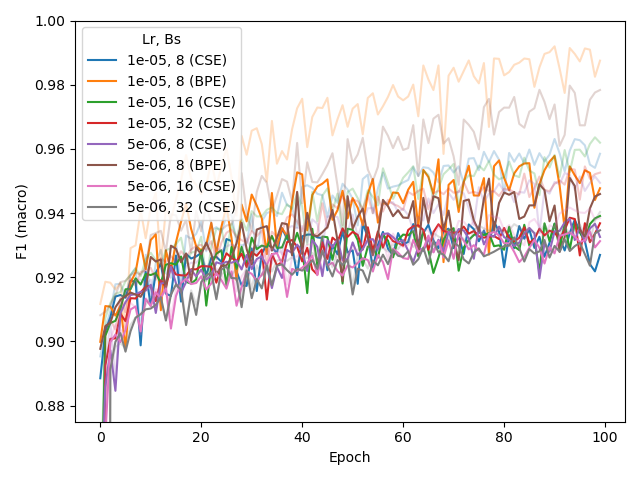
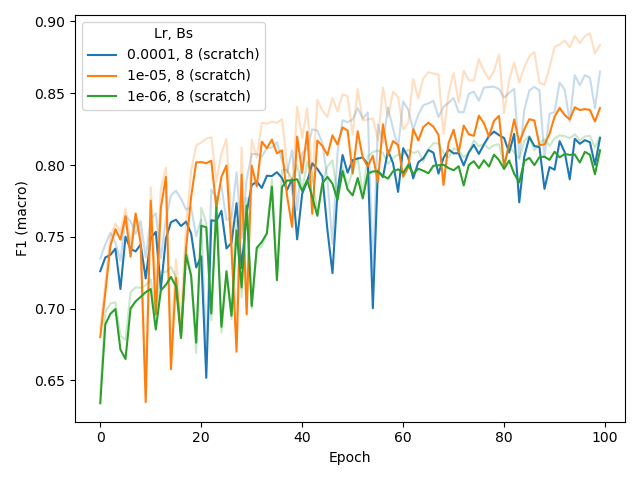
appropriate for BERT-medium in combination with BPE or CSE (below, middle), also when training from scratch (instead of fine-tuning a pre-trained model, for CSE, below, right).
This more recent experiment also included several batch sizes (bs), for which batch_size=8 was found to be best (also identified at an earlier stage but not reported here).
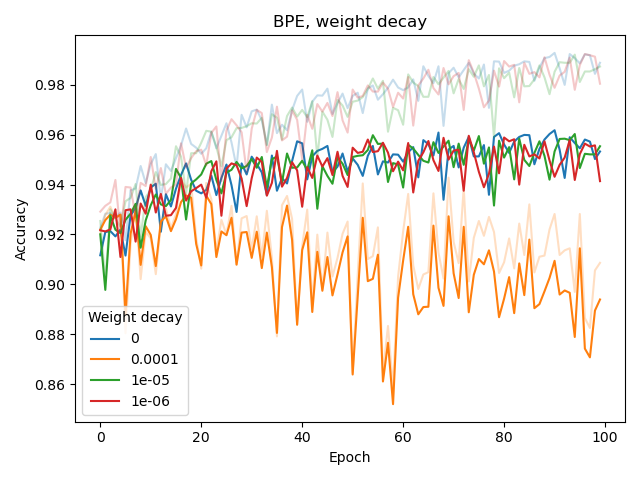
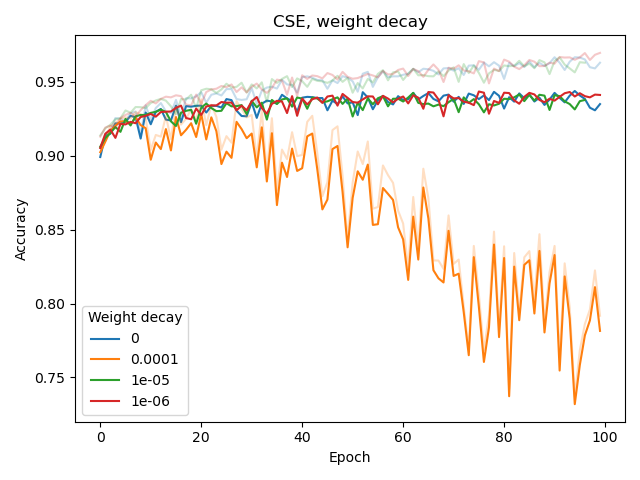
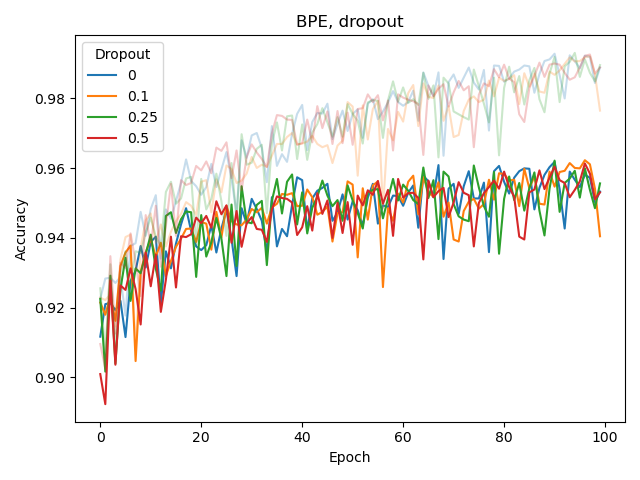
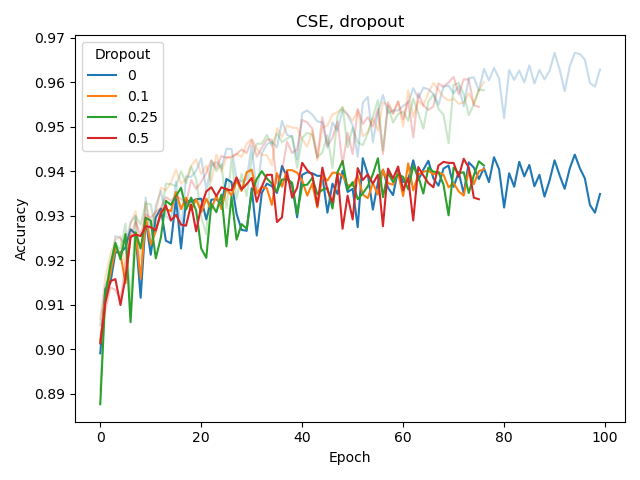
We also experimented with the weight_decay hyperparameter of Adam as well as adding a dropout layer before the classification (sigmoid) output layer, both for BPE and CSE.
None of these seemed to improve classification or reduce overfitting. The latter (overfitting) was assessed by considering the difference between training and validation performance, with training performance plotted as a lighter-colored curve.
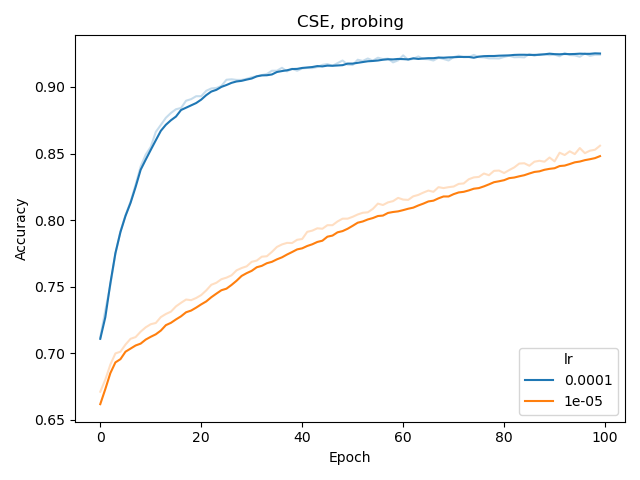
Finally, for the probing the network with a tiny MLP (256 hidden nodes), we found that a learning rate of 0.0001 was more appropriate.
Convolutional Sequence Encoding
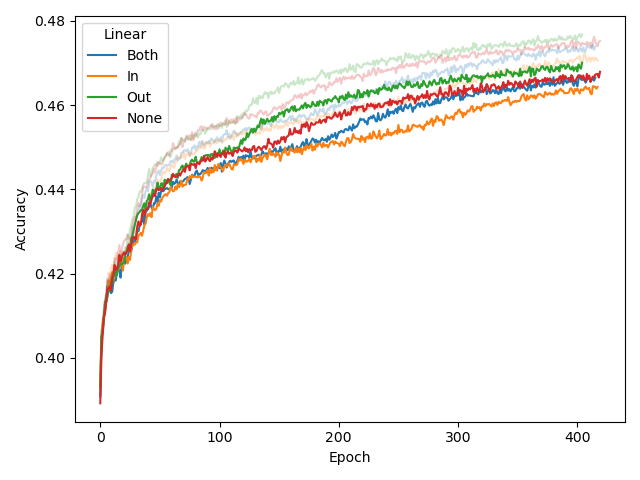
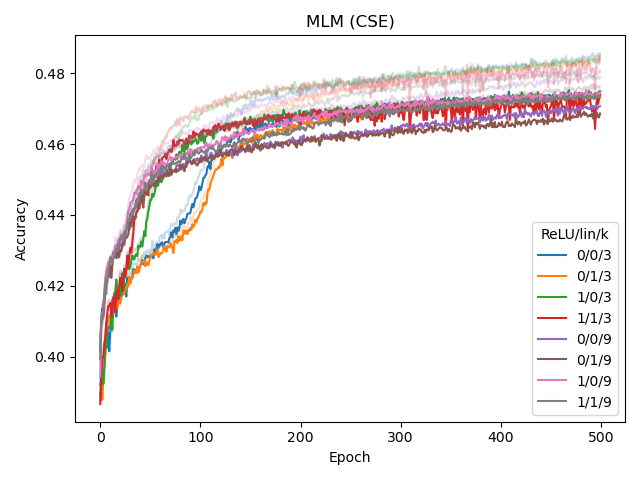
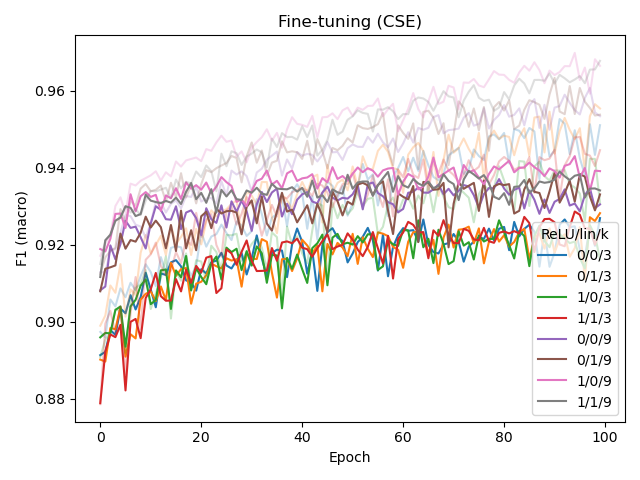
We experimented with several architectural changes for the CSE component during its development, while using BERT-medium as base architecture. Based on the left plot below, we conclude that adding a linear layer before the final MLM output layer helps the model, while adding an extra linear (embedding) layer after the convolution in the input does not. The input CSE layer is further experimented with in the middle and right plot, which shows that ReLU-activating the convolution (but not linearly transforming it) helps for CSE with larger k (in this case, 9).
While linearly transforming the convolved embeddings in the input layer was shown to have a negative affect, it did allow us to set the number of kernels to a value other than d_model.
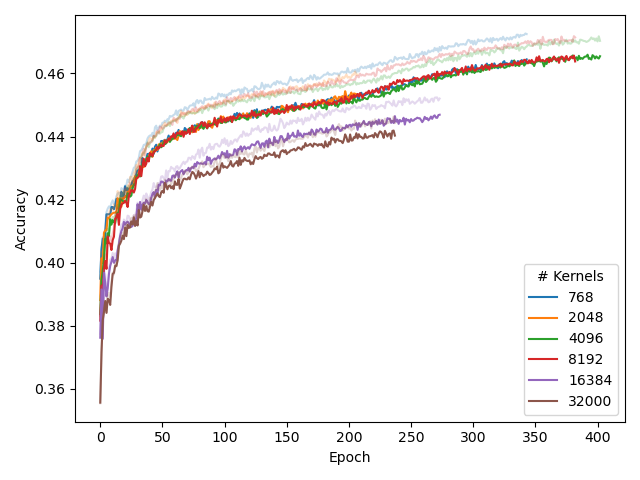
Hence, we experimented with several different number of kernels in the CSE, and then transforming the output to d_model dimensions with a linear layer.
Our results (left plot) indicate that adding more kernels does not benefit performance. Adding too many kernels can even deteroriate performance.
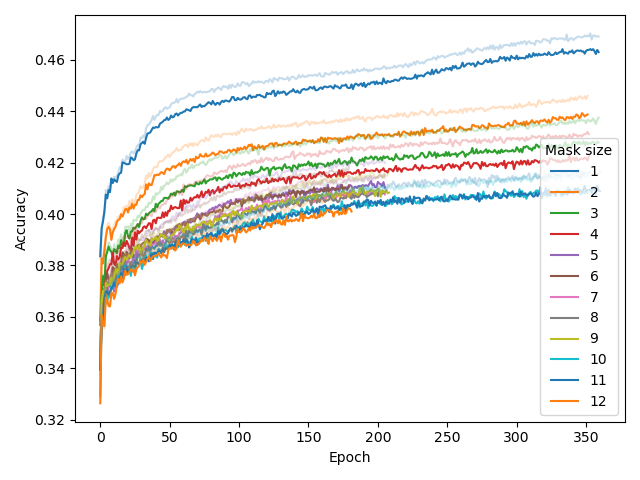
We also experimented whether masking out multiple consecutive nucleotides (defined by the mask_size hyperparameter, similar to SPAN-BERT) would improve pre-training (middle plot). This was not the case. In fact, we found that a model pre-trained with a mask size of 1 would perform better at predicting multiple masked nucleotides than a model pre-trained with mask size of 2.
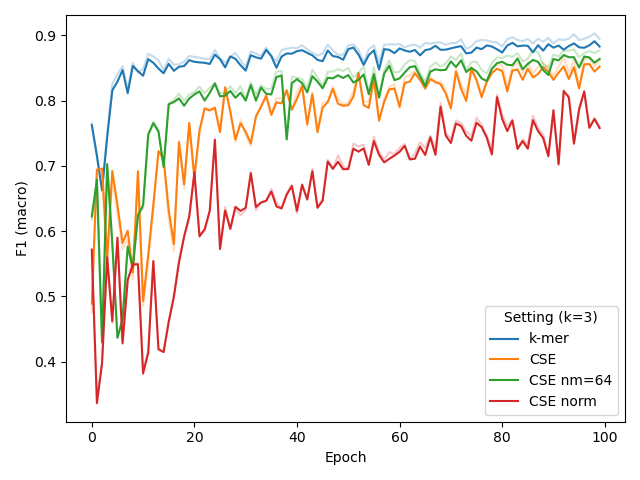
In a final attempt to increase CSE’s performance for short k, (i.e. 3), we tried adding layer normalization after convolution or significantly reducing the number of kernels to 64, but this did not help (right plot).